About
News
1/3/24: Thejesh joins the lab!
Thejesh Mallidi got his Bachelor’s degree in Computer Science and is now a Master’s student in Data Science at MSU. He’s spent the last two years honing his skills as a Machine Learning Engineer specializing in Applied Machine Learning, Computer Vision, Deep Learning and Natural Language Processing. His background includes hands-on experience in developing and deploying machine learning models for various applications. His current research focus is on applying large language models to plant science literature. Welcome, Thejesh!
9/1/23: Our newest PhD sudent, Jingyao!
Welcome to the lab, Jingyao! Jingyao Tang is a PhD student in the Department of Computational Mathematics, Science, and Engineering. She studied Applied Statistics for her Master’s degree while at Syracuse Univeristy in New York.
8/17/23: Congratulations to Brianna on becoming a PhD candidate!
Brianna presented her research on utilizing machine learning and AI techniques to elucidate genetic mechanisms important under abiotic stress. We are very proud of her!
3/15/23: Welcome, Paulo!
Dr. Paulo Izquierdo has joined the lab as a Great Lakes Bioenergy Research Center postdoctoral research associate, having recently completed his PhD in Plant Breeding, Genetics, and Biotechnology here at Michigan State. During his PhD, he worked on implementing genomic prediction methods for research on dry beans to increase the genetic gain per unit of time in traits such as yield, mineral concentration, and end-use quality traits. His research focus now is on plant computational sciences and genomic prediction in switchgrass. We are glad to have you!
2/1/23: Welcome, Rajneesh!
Welcome Dr. Rajneesh Singhal who has decided to join the lab as a postdoctoral research associate!
10/10/22: Thilanka’s study on switchgrass transcriptional cold-response is published in Frontiers in Plant Science! Congratulations!
In collaboration with several members in the lab (Brianna, Peipei, and Shinhan), they investigated the Temporal regulation of of cold transcriptional response in switchgrass. Switchgrass low-land ecotypes have significantly more biomass but lower cold tolerance than up-land ecotypes. Therefore, understanding the molecular mechanisms underlying cold response in up-land ecotypes can contribute to improving cold tolerance of high-yielding low-land ecotypes under chilling and freezing environmental conditions. They dissect the cis-regulatory basis of switchgrass cold stress transcriptional response using machine learning methods.
10/5/22: 2 new members join the lab! Welcome, Edmond and Mae!
Edmond Anderson rotated in the lab for 1 month and he has decided to join us for the remainder of his masters program. He is studying data science. Mae Milton is a third year undergraduate student. We’re happy to have you both!
8/31/22: Ally becomes a PhD candidate!
Congratulations, Ally! Ally’s presentation was on predicting co-functional gene pairs in Arabidopsis and yeast.
8/8/22: Welcome to the lab Isaia!
Isaia is an exchange graduate student from the Heinrich-Heine University of Düsseldorf in Düsseldorf, Germany. We are very excited to have him here with us. Welcome to MSU!
8/1/22: Eleanor joins the Shiu Lab!
 Eleanor initially joined the lab in June as an intern from the University of Michigan’s Data Science Master’s Program, and has now decided to stay with us! She will be working as a research technician in our lab. We are happy to have you, Eleanor!
Eleanor initially joined the lab in June as an intern from the University of Michigan’s Data Science Master’s Program, and has now decided to stay with us! She will be working as a research technician in our lab. We are happy to have you, Eleanor!
5/26/22: Kenia becomes a PhD candidate!
Kenia presented an hour-long seminar on her proposed thesis research. Her talk was about dissecting changes in the genetic architecture of fitness across environments and cross-species predictions of genetic interactions.
4/26/22: A big congratulations to Elijah, who was accepted into an REU program!
Elijah will spend the summer at Washington State University where he will be working with Dr. Arron H. Carter’s lab as part of the USDA/NIFA FACT: Research Experience for Undergraduates on Phenomics Big Data Management program.
3/16/22: We welcome Krishen to the lab!
 Krishen is a third year undergraduate student studying Plant Biology, Biochemistry, Human Biology, and Genetics. We’re so happy to have you here with us!
Krishen is a third year undergraduate student studying Plant Biology, Biochemistry, Human Biology, and Genetics. We’re so happy to have you here with us!
2/21/22: Serena interviewed about her research for the Sci-Files podcast
 Sci-Files is a science communication podcast produced by MSU’s SciComm club, where graduate students are interviewed about their research. Serena was interviewed about her work on knowledge graphs for the February 20th episode, which is published here as a podcast.
Sci-Files is a science communication podcast produced by MSU’s SciComm club, where graduate students are interviewed about their research. Serena was interviewed about her work on knowledge graphs for the February 20th episode, which is published here as a podcast.
2/9/22: Peipei and Fanrui’s study on measuring Arabidopsis fitness using Faster R-CNN is accepted
Fitness (number of seeds or fruits producted by an individual) is challenging to measure accurately due to the high fecundity and relatively small propagule sizes. Peipei and Fanrui, with significant contribution from other colleagues, established Faster R-CNN based object detection models which can automatively detect and count Arabidopsis seeds and siliques in high throughput. This work is now accepted in New Phytologist.
1/22/22: Peipei’s review paper on computational prediction of plant metabolic pathway genes is published
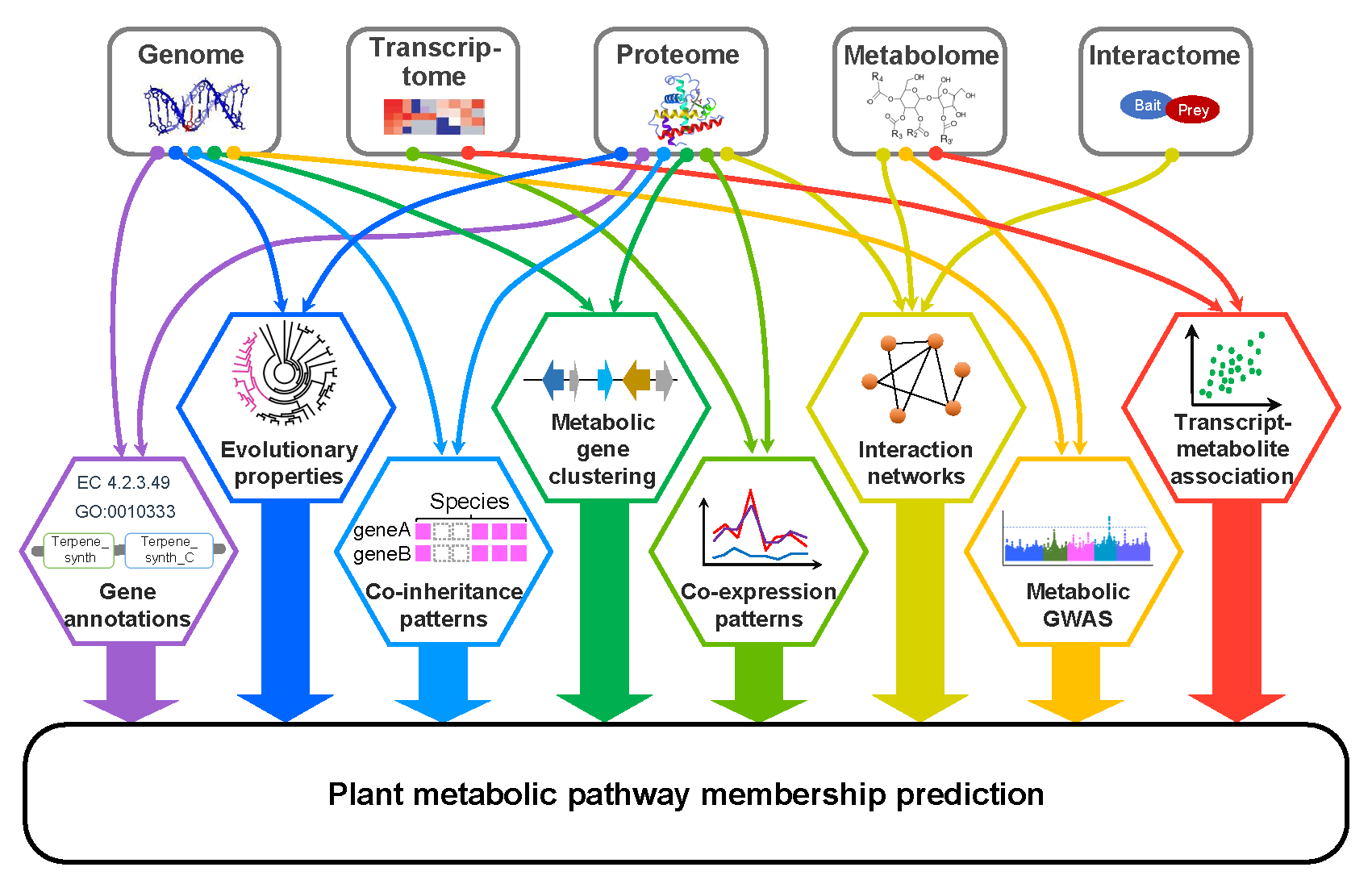 Uncovering genes encoding enzymes responsible for the biosynthesis of diverse plant metabolites is essential for metabolic engineering and production of plant metabolite-derived medicine. Peipei, Ally and Shin-Han wrote a review paper regarding recent progress in the computational prediction of plant metabolic pathway genes. This review is now published in Current Opinion in Plant Biology.
Uncovering genes encoding enzymes responsible for the biosynthesis of diverse plant metabolites is essential for metabolic engineering and production of plant metabolite-derived medicine. Peipei, Ally and Shin-Han wrote a review paper regarding recent progress in the computational prediction of plant metabolic pathway genes. This review is now published in Current Opinion in Plant Biology.
12/2/21: Beth’s study on wounding transcriptional responses is published
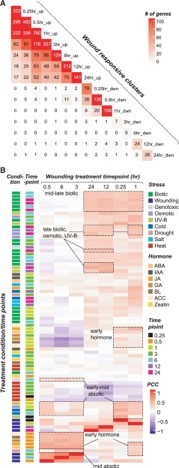 Plants respond to wounding stress by changing gene expression patterns and inducing the production of hormones including jasmonic acid. Bethany led a study modeling the temporal and hormonal regulation of plant transcriptional response to wounding in Arabidopsis. This work is now published in The Plant Cell.
Plants respond to wounding stress by changing gene expression patterns and inducing the production of hormones including jasmonic acid. Bethany led a study modeling the temporal and hormonal regulation of plant transcriptional response to wounding in Arabidopsis. This work is now published in The Plant Cell.
10/12/21: Welcome to the lab, Huan!
 Huan is officially a member of the Shiu lab! She is a third year graduate student in the Genetics and Genome Sciences and Molecular Plant Sciences Programs. We’re glad to have you Huan!
Huan is officially a member of the Shiu lab! She is a third year graduate student in the Genetics and Genome Sciences and Molecular Plant Sciences Programs. We’re glad to have you Huan!
10/5/21: Welcome aboard, Elijah!
 We are very excited to welcome Elijah to the team! Elijah is a third year undergraduate student majoring in Plant Biology.
We are very excited to welcome Elijah to the team! Elijah is a third year undergraduate student majoring in Plant Biology.
09/21/21: Brianna joins the lab!
 Brianna rotated in the lab during the summer and surprised us all one day when she walked in and exclaimed “I’m joining the lab!”. She graduated from Stanford University with a Bachelor’s degree in Earth Systems and Human Biology. Brianna is also an NRT-IMPACTS fellow and is a first year graduate student in the Plant Biology program. Welcome Brianna!
Brianna rotated in the lab during the summer and surprised us all one day when she walked in and exclaimed “I’m joining the lab!”. She graduated from Stanford University with a Bachelor’s degree in Earth Systems and Human Biology. Brianna is also an NRT-IMPACTS fellow and is a first year graduate student in the Plant Biology program. Welcome Brianna!
8/1/21: New grant for the lab

7/29/21: Siobhan’s study on modeling plant genetic redundancy is published
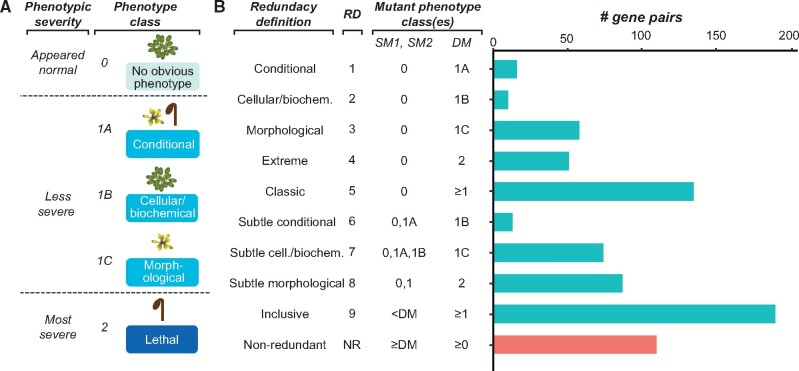 Genetic redundancy refers to a situation where an individual with a loss-of-function mutation in one gene (single mutant) does not show an apparent phenotype until one or more paralogs are also knocked out (double/higher-order mutant). Siobhan led a study aiming at understanding the factors contributing to genetic redundancy with machine learning approaches. In addition to Siobhan, Peipei, Serena, Beth, Fanrui, and Melissa have contributed significantly to the study. This work is now published in Molecular Biology & Evolution.
Genetic redundancy refers to a situation where an individual with a loss-of-function mutation in one gene (single mutant) does not show an apparent phenotype until one or more paralogs are also knocked out (double/higher-order mutant). Siobhan led a study aiming at understanding the factors contributing to genetic redundancy with machine learning approaches. In addition to Siobhan, Peipei, Serena, Beth, Fanrui, and Melissa have contributed significantly to the study. This work is now published in Molecular Biology & Evolution.
7/20/21: An essay on overcoming challenges to enhancing experimental biology with computational modeling is published
 Computational modeling approaches are underutilized in plant biology. Lead by Renee Dale from Danforth Plant Science Center, Shinhan and seven other colleagues published an opinion piece in Frontiers in Plant Science identifying challenges one may face in adopting modeling approach.
Computational modeling approaches are underutilized in plant biology. Lead by Renee Dale from Danforth Plant Science Center, Shinhan and seven other colleagues published an opinion piece in Frontiers in Plant Science identifying challenges one may face in adopting modeling approach.
7/5/21: The Lab RETURNS!
For the past school year 2020-2021 the lab has been working remotely. Today was the first time all members were able to experience the lab environment together in over a year! This was Kenia and Ally’s first oppertunity to settle into the lab space due to their first year being entirely remote.
6/2/21: Kenia is awarded NRT-IMPACTS and PBHS fellowships
Congratulations to Kenia for receiving an NRT-IMPACTS fellowship, a training program for doctoral students to apply computational approaches to address challenges in plant biology. Kenia also received the PBHS fellowship, which is an NIH funded T32 training program for predoctoral students to pursue research related to plant biotechnology and chemical engineering.
6/1/21: Peipei’s study on metabolic pathway membership prediction is published
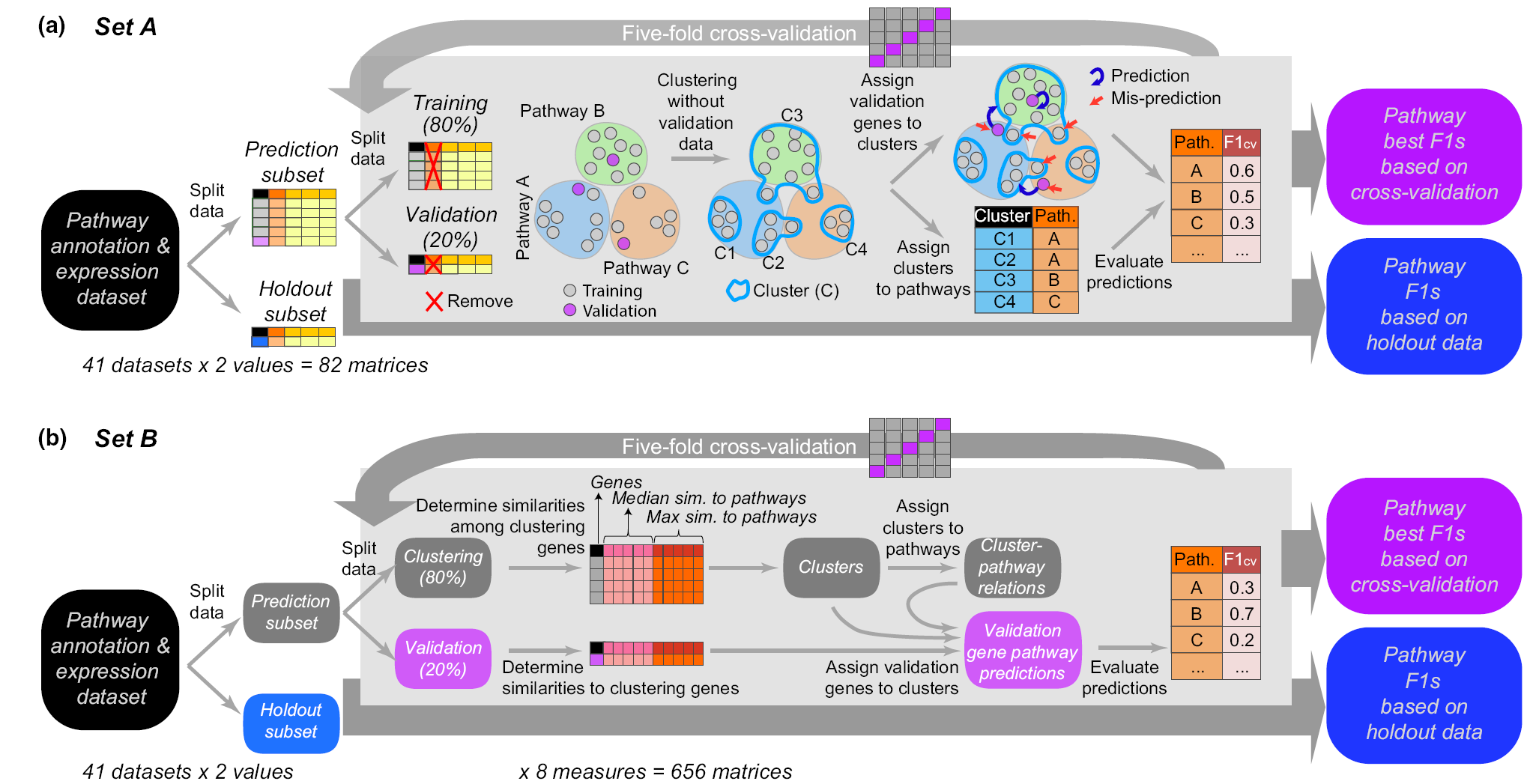 Genes in the same metabolic pathway are assumed to be co-expressed. This study, led by Peipei, aiming to use multiple strategies (including unsupervised and supervised machine learning) to evaluated the utility of gene expression data in the prediction of plant metabolic pathway membership. In addition to Peipei, Bethany, Sahra, Melissa, and Cornelius have contributed significantly to the study. This work is now published in New Phytologist.
Genes in the same metabolic pathway are assumed to be co-expressed. This study, led by Peipei, aiming to use multiple strategies (including unsupervised and supervised machine learning) to evaluated the utility of gene expression data in the prediction of plant metabolic pathway membership. In addition to Peipei, Bethany, Sahra, Melissa, and Cornelius have contributed significantly to the study. This work is now published in New Phytologist.
5/11/21: Serena presents an introduction to machine learning at the GLBRC Annual Science Meeting
 Machine learning (ML) is an important tool for making and interpreting predictions in biology. Serena gave a talk introducing the basic concepts of ML and how it can be used to answer biological questions at GLBRC’s Annual Science Meeting. A recording of the presentation can be found here
Machine learning (ML) is an important tool for making and interpreting predictions in biology. Serena gave a talk introducing the basic concepts of ML and how it can be used to answer biological questions at GLBRC’s Annual Science Meeting. A recording of the presentation can be found here
5/10/21: Serena becomes a PhD candidate
 Serena gave a one-hour seminar on her proposed thesis research as part of the second half of her comprehensive exam. Her talk was about automated information extraction and knowledge graphs, and how we can apply them to generate novel hypotheses in the plant sciences. Afterwards she met with her committee for the proposal defense, and passed!
Serena gave a one-hour seminar on her proposed thesis research as part of the second half of her comprehensive exam. Her talk was about automated information extraction and knowledge graphs, and how we can apply them to generate novel hypotheses in the plant sciences. Afterwards she met with her committee for the proposal defense, and passed!
4/17/21: Why are there no symptoms when a plant is infected with a fungal pathogen?
 The broad host range of Fusarium virguliforme represents a unique comparative system to identify and define differentially induced responses between an asymptomatic monocot host, maize (Zea mays), and a symptomatic eudicot host, soybean (Glycine max). Led by Amy Baetsen-Young from Brad Day’s lab, Shinhan collaborated on a project examining how asymptomatic and symptomatic hosts respond to F. virguliforme transcriptionally. This work is now published in Plant Cell
The broad host range of Fusarium virguliforme represents a unique comparative system to identify and define differentially induced responses between an asymptomatic monocot host, maize (Zea mays), and a symptomatic eudicot host, soybean (Glycine max). Led by Amy Baetsen-Young from Brad Day’s lab, Shinhan collaborated on a project examining how asymptomatic and symptomatic hosts respond to F. virguliforme transcriptionally. This work is now published in Plant Cell
2/2/21: Peipei’s study on genome misassembly is published
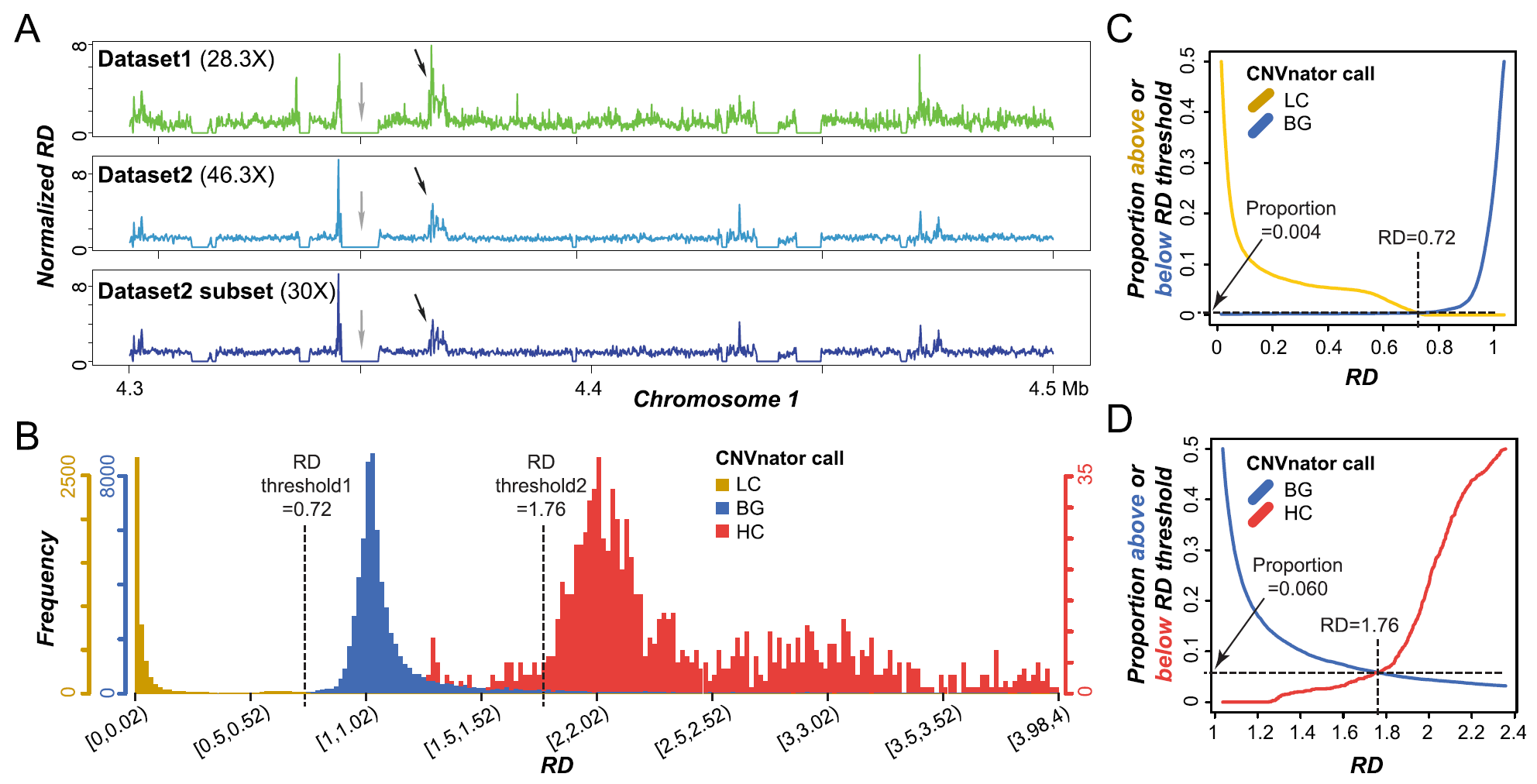 Tandemly duplicated genomic regions can be misassembled together due to high sequence similarity in short read-based plant assembly, resulting higher read coverages than other regions. Peipei, Fanrui, and Bethany used comparative genomics and machine learning approaches to investigate the factors contributing to the uneven distribution of read depth and the potential misassembly. This work is now published in BMC Genomics.
Tandemly duplicated genomic regions can be misassembled together due to high sequence similarity in short read-based plant assembly, resulting higher read coverages than other regions. Peipei, Fanrui, and Bethany used comparative genomics and machine learning approaches to investigate the factors contributing to the uneven distribution of read depth and the potential misassembly. This work is now published in BMC Genomics.
12/18/20: Lab member Abigail Seeger completed her undergraduate career
 Abigail graduated from Lyman Briggs College with degrees in Statistics and Plant Biology in a virtual commencement ceremony. She plans to continue working with the lab in the spring ‘21.
Abigail graduated from Lyman Briggs College with degrees in Statistics and Plant Biology in a virtual commencement ceremony. She plans to continue working with the lab in the spring ‘21.
12/7/20: Welcome the newest member of our lab - Kenia Segura Abá
 Kenia is a first year graduate student in the Genetics and Genome Sciences Program. She rotated over the summer and has made the decision to join our lab today! She graduated from University of Texas-Austin with families in the San Antonio area. Welcome to our lab Kenia!
Kenia is a first year graduate student in the Genetics and Genome Sciences Program. She rotated over the summer and has made the decision to join our lab today! She graduated from University of Texas-Austin with families in the San Antonio area. Welcome to our lab Kenia!
12/3/20: Serena’s paper is published with gazillion authors
The paper that Serena worked on during the IMPACTS course Foundations in Computational Plant Science, Composite modeling of leaf shape along shoots discriminates Vitis species better than individual leaves, has been published! A collaborative effort amont more than 30 authors, the paper is in the December issue of the journal Applications in Plant Sciences
9/21/20: Serena gave lesson in IMPACTS class
Today during the Foundations of Computational Plant Sciences course, Serena and the team of IMPACTS trainees from the Spring 2020 Forum in Computational Plant Sciences presented their lesson plan to students. This lesson plan will soon be submitted for publication in the journal CourseSource.
9/1/20: Collaborative work on maize ANT1 regulatory network is published!
Arabidopsis AINTEGUMENTA (ANT), an AP2 transcription factor, is known to control plant growth and floral organogenesis. This study is led by Dr. Wen-Hsiung Li from Academia Sinica, Taiwan, which reveals biological roles of ANT1 in several developmental processes beyond its known roles in plant growth and floral organogenesis. It is published in the Sep. 1st issue of Proc. Natl. Acad. Sci., USA.
9/1/20: Christina’s story on predictions of plant stress response through multi-omics data integration is published!
Plants respond to their environment by dynamically modulating gene expression. Our findings demonstrate how in silico approaches can improve our understanding of the complex codes regulating response to combined stress and help us identify prime targets for future characterization. It is published in Nuc. Acid. Res. Genomics and Bioinformatics.
7/30/20: Beth’s story on using Arabidopsis annotation information to improve predictions of tomato metabolic genes is published!
Plant specialized metabolites mediate interactions between plants and the environment and have significant agronomical/pharmaceutical value. This study demonstrates that specialized and general metabolic genes can be better predicted by leveraging cross-species information. Additionally, our findings provide an example for transfer learning in genomics where knowledge can be transferred from an information-rich species to an information-poor one. The findings is now available form In Silico Plant.
7/2/20: Collaborative work on the evolution of a gene cluster in Solanaceae is published!
The hairs on the surface of some species in the nightshade family (Solanaceae) produce acylsugars which deter herbivores and pests from damaging the plants. A tomato gene cluster involved in medium chain acylsugar accumulation was described, and the evolution of this gene cluster was reconstructed. This study is led by Dr. Pengxiang Fan from Prof. Robert L Last’s lab, and Peipei has contributed significantly to the evolutionary history reconstruction. It is published in Elife.
7/1/20: Ally’s offically first day!
Ally has known she was joing the lab and has been occationally joing the lab for lunch meetings up to this point. Now it is the first day as an offical graduate student here at MSU for Ally. Welcoming her to the lab with virtual open arms!
6/25/20: Serena won!!
 Serena is the winner of the SciComm Blog Contest! Here is her winning post KNOWLEDGE GRAPHS: A New Way To Reason. Congrats Serena!
Serena is the winner of the SciComm Blog Contest! Here is her winning post KNOWLEDGE GRAPHS: A New Way To Reason. Congrats Serena!
6/21/20: Virtual lunch in its 3rd month… Now with Fanrui!!
 Fanrui, our deep learning expert, Peipei’s beloved husband, and a great cook, is finally back after getting stuck in China for >6 months due to the COVID19, repeated flight canceldation, and long stay in the secondary screen through the immigration. Welcome back!
Fanrui, our deep learning expert, Peipei’s beloved husband, and a great cook, is finally back after getting stuck in China for >6 months due to the COVID19, repeated flight canceldation, and long stay in the secondary screen through the immigration. Welcome back!
5/13/20: Virtual lab lunch at its second month
 In our lab lunch today, Ronan is biting her mom’s face!
In our lab lunch today, Ronan is biting her mom’s face!
5/11/20: Christina’s review on interpretable machine learning is published
 In collaboration with Jiliang Tang from Comp. Sci. & Engr, MSU, Christina’s review on Opening the Black Box: Interpretable Machine Learning for Geneticists is now published in Trends in Genetics. This work highlights current progress in making machine learning model interpretable and explanable using applications in genetics as examples. And her graphics was picked as the cover as shown on the right!
In collaboration with Jiliang Tang from Comp. Sci. & Engr, MSU, Christina’s review on Opening the Black Box: Interpretable Machine Learning for Geneticists is now published in Trends in Genetics. This work highlights current progress in making machine learning model interpretable and explanable using applications in genetics as examples. And her graphics was picked as the cover as shown on the right!
4/8/20: Virtual lab lunch for almost a month
 With the social distancing in place, we continue to meet over Zoom. It seems that folks are doing ok!
With the social distancing in place, we continue to meet over Zoom. It seems that folks are doing ok!
3/31/20: Collaborative work with Dr. Yan Bao is published
Dr. Bao’s work on COST1 regulates autophagy to control plant drought tolerance is published where Peipei has contributed significantly using comparative genomics approaches.
3/19/20: Siobhan defended today
 Siobahn defended her thesis titled Modeling and prediction of genetic redundancy in Arabidopsis thaliana and Saccharomyces cerevisiae today virtually via Zoom with 89 people in the audience! Both us showed up in the lab (while keeping >6 ft distance) to get the virtual defense setup. Originally due to move to England next week, Siobhan will stay in East Lansing to finish her work. Good job Siobhan! The picture on the right shows Siobhan right after the defense.
Siobahn defended her thesis titled Modeling and prediction of genetic redundancy in Arabidopsis thaliana and Saccharomyces cerevisiae today virtually via Zoom with 89 people in the audience! Both us showed up in the lab (while keeping >6 ft distance) to get the virtual defense setup. Originally due to move to England next week, Siobhan will stay in East Lansing to finish her work. Good job Siobhan! The picture on the right shows Siobhan right after the defense.
3/13/20: Our first virtual lab lunch
 With all of us working remotely, we decide to host a 15-min lab lunch to check in with everyone. The photo on the right is all of us experimenting with the background feature in Zoom - the two “ghosts” are Siobhan and Serena!
With all of us working remotely, we decide to host a 15-min lab lunch to check in with everyone. The photo on the right is all of us experimenting with the background feature in Zoom - the two “ghosts” are Siobhan and Serena!
3/11/20: Social distancing starts
Michigan State has indicated all classes will be online only today. Given the nature of most of our work is computational and considering the risks of COVID-19, from today on all lab personnel will be working remotely, except for those who need to keep our plant alive.
3/5/20: Welcome to the lab Ally
 Ally has decided to join the Plant Biology Program and, instead of doing any rotation, is coming to our lab! She is currently finishing up her Bachelor’s degree in Manchester University, a liberal art institution. She is awarded an MSU College of Natural Science Fellowship as well as an Early-Start Fellowship to begin her doctoral training on July 1st! Welcome to our lab Ally!
Ally has decided to join the Plant Biology Program and, instead of doing any rotation, is coming to our lab! She is currently finishing up her Bachelor’s degree in Manchester University, a liberal art institution. She is awarded an MSU College of Natural Science Fellowship as well as an Early-Start Fellowship to begin her doctoral training on July 1st! Welcome to our lab Ally!
2/21/20: Welcome to the lab Serena
 Serena is a new graduate student in the, hold your breath, Plant Biology Program, the Computational Math., Sci., Math Program, AND the Moleculr Plant Science Program. She is a MSU Plant Science Fellow and an NSF Fellow of the IMPACTS training program. She graduated from the Cornell University in 2019 and has been rotating in two other labs. Here is the funny Slack exchange when I found out she is joining us!
Serena is a new graduate student in the, hold your breath, Plant Biology Program, the Computational Math., Sci., Math Program, AND the Moleculr Plant Science Program. She is a MSU Plant Science Fellow and an NSF Fellow of the IMPACTS training program. She graduated from the Cornell University in 2019 and has been rotating in two other labs. Here is the funny Slack exchange when I found out she is joining us!
1/1/20: Christina’s study on using genomic and transcriptomic information to predict trait is published
Can transcriptome data be used to predict traits? In the study titled Transcriptome-Based Prediction of Complex Traits in Maize, Christina, in collaboration with Jeremy, a rotating student at the time, and Bob VanBuren tested this and found out that not only we can predict traits with transcriptomes, but they may tell us more than genomic data can. The finding is featured in MSU Today!
10/25/19: Beth defended her PhD
 Beth defended her PhD thesis today titled Specilized metabolism and stress response: studies in predicting gene function and regulation. The seminar was held in Plant Biology Lab 247 and was attended by both her colleagues, friends, and families. Beth is moving to University of Wisconsin-Madison to take a
Beth defended her PhD thesis today titled Specilized metabolism and stress response: studies in predicting gene function and regulation. The seminar was held in Plant Biology Lab 247 and was attended by both her colleagues, friends, and families. Beth is moving to University of Wisconsin-Madison to take a
toral position applying molecular evolutionary and computaional approaches to plant biochemistry. We wish her all the best! The picture on the right shows Beth right before her defense.
10/20/19: Ronan is here!
Siobhan gave birth today to her baby boy Ronan! Congrats Siobhan and Geoff!
9/27/19: Christina defeneded her PhD
 Christina today defended her PhD thesis titled Interpretable machine laerning in plant genomes: studies in the complex relationship between genotype and phenotype. The seminar was held in Plant Biology Lab 247 and was full! Christina’s parents also attended her seminar. In the talk, Christina laid out the overarching Christina is now moving to Australia working in algorithm development in single cell genomics studies. All the best Christina! The picture on the right showing Chrisitna leaving the lab for good on 10/3/2020…
Christina today defended her PhD thesis titled Interpretable machine laerning in plant genomes: studies in the complex relationship between genotype and phenotype. The seminar was held in Plant Biology Lab 247 and was full! Christina’s parents also attended her seminar. In the talk, Christina laid out the overarching Christina is now moving to Australia working in algorithm development in single cell genomics studies. All the best Christina! The picture on the right showing Chrisitna leaving the lab for good on 10/3/2020…
9/24/19: Sahra and Christina’s work on the regulatory mechanisms of cell-type stress response is published
Our story titled Cis-regulatory code for predicting plant cell-type transcriptional response to high salinity is now published in Plant Physiology with Sahra and Christina as joint first authors. Using the root cell-type transcriptome data, Sahra and Christina identified cis-regulatory sequences likely specify cell-type response to high salinity stress. More importantly, machine learning models were built to ask how well the identified regulatory sequences can predict cell-type response. The findings not only advance our understanding of the regulatory mechanisms of the plant spatial transcriptional response, but also suggest broad applicability of the approach to any species, particularly those with little or no trans regulatory data.
9/18/19: Christina’s work on benchmarking genomic prediction algorithms is published
Chrisitina’s paper on Benchmarking Parametric and Machine Learning Models for Genomic Prediction of Complex Traits is published today in G3. This is a collaborative between our colleague Gustavo de los Compos, Chrisitna’s internship mentors Andrew McCarren and Mark Roantree, and Emily Bolger a highly productive summer research student. Using data of 18 traits across six plant species with different marker densities and training population sizes, Christina spearheaded a comparison of the performance of six linear and six non-linear algorithms. The findings highlight that simpler, linear algorithm can beat out more sophiscated machine learning approaches, and the importance of algorithm selection for the prediction of trait values.
8/20/19: John’s work on poaceae intergenic transcription is published
John’s paper on Evolutionary characteristics of intergenic transcribed regions indicate rare novel genes and widespread noisy transcription in the Poaceae is published in Scientific Report. Christina as well as Rosalie Sowers, a highly talented undergrad from Penn State also contribute to this work. Extensive transcriptional activity occurring in intergenic regions of genomes has raised the question whether intergenic transcription represents the activity of novel genes or noisy expression. To assess this, John et al. evaluated cross-species and post-duplication sequence and expression conservation of intergenic transcribed regions. This study provides a framework to identify novel genes using comparative transcriptomic data to improve genome annotation that is fundamental for connecting genotype to phenotype in crop and model systems.
8/19/19: Our newest addition to the lab, Thilanka
 Thilanka is a new graduate student in the Plant Biology Graduate Program. He graduated from the University of Peradeniya in Sri Lanka in 2016 and worked as a teaching assistant in the past two years in Peradeniya. He is energetic, communicative, and excited about learning new topics. He is also easy to get alone with, and has a black belt in Taekwondo!! Welcome to the lab!
Thilanka is a new graduate student in the Plant Biology Graduate Program. He graduated from the University of Peradeniya in Sri Lanka in 2016 and worked as a teaching assistant in the past two years in Peradeniya. He is energetic, communicative, and excited about learning new topics. He is also easy to get alone with, and has a black belt in Taekwondo!! Welcome to the lab!
7/24/19: Siobhan got the Dissertation Completion Fellowship
Siobhan has been selected to receive a $7,500 College of Natural Science Dissertation Completion Fellowship during fall semester 2019. Congrats Siobhan!
7/13/19: Nick’s work on regulatory assymmetry of plant duplicate genes is published
Nick’s paper on Expression and regulatory asymmetry of retained Arabidopsis thaliana transcription factor genes derived from whole genome duplication is published in BMC Evoltionary Biology. Christina and Eamon Winship, an undergraduate reseracher at the time also contributed. Transcription factors (TFs) play a key role in gene regulation and tend to be retained after duplication. Nick’s work provide answers about about how TF duplicates have diverged in their expression and regulation that contribute to a better understanding of the elevated retention rate among TFs.
2/19/19: Our collaborative work on maize time-series transcriptome is published
Led by Dr. Wen-Hsiung Li’s group in Taiwan, Shinhan has contributed to the study: Comparative transcriptomics method to infer gene coexpression networks and its applications to maize and rice leaf transcriptomes published in PNAS. This study involves a comprehensive time series data and a novel approach to build time-resolved co-expression network to hypothesize regulatory relationships between genes.
2/5/19: Beth’s work on specialized metabolism is published and featured by NSF
Beth’s work on Robust predictions of specialized metabolism genes through machine learning is published in PNAS with Peipei, Johnny, Melissa, and multiple colleagues in Rob Last’s group as co-authors. This work features a machine learning approach to predict general and specialized metabolic gene that are notoriously hard to distinquish. The work is featured in MSU Today and in NSF’s Awesome Discoveries You Probably Didn’t Hear About This Week.
